Incoherence is not ideal, but it is a realistic part of any work such as we are doing within the openEHR community. Transparency and openness can mitigate some of the incoherence. Within a transparent, governed and collaborative environment incoherence and apparent conflict can be recognised and leveraged constructively to improve the quality of archetypes.
Archetype Re-use
Last Thursday & Friday @hughleslie and I presented a two day training course on openEHR clinical modelling. Introductory training typically starts with a day to provide an overview – the "what, why, how" about openEHR, a demo of the clinical modelling methodology and tooling, followed by setting the context about where and how it is being used around the world. Day Two is usually aimed at putting away the theoretical powerpoints and getting everyone involved - hands on modelling. At the end of Day One I asked the trainees to select something they will need to model in coming months and set it as our challenge for the next day. We talked about the possibility health or discharge summaries – that's pretty easy as we largely have the quite mature content for these and other continuity of care documents. What they actually sent through was an Antineoplastic Drug Administration Checklist, a Chemotherapy Ambulatory Care Nursing Intervention and Antineoplastic Drug Patient Assessment Tool! Sounded rather daunting! Although all very relevant to this group and the content they will have to create for the new oncology EHR they are building.
Perusing the Drug Checklist ifrst - it was easily apparent it going to need template comprising mostly ACTION archetypes but it meant starting with some fairly advanced modelling which wasn't the intent as an initial learning exercise.. The Patient Assessment Tool, primarily a checklist, had its own tricky issues about what to persist sensibly in an EHR. So we decided to leave these two for Day Three or Four or..!
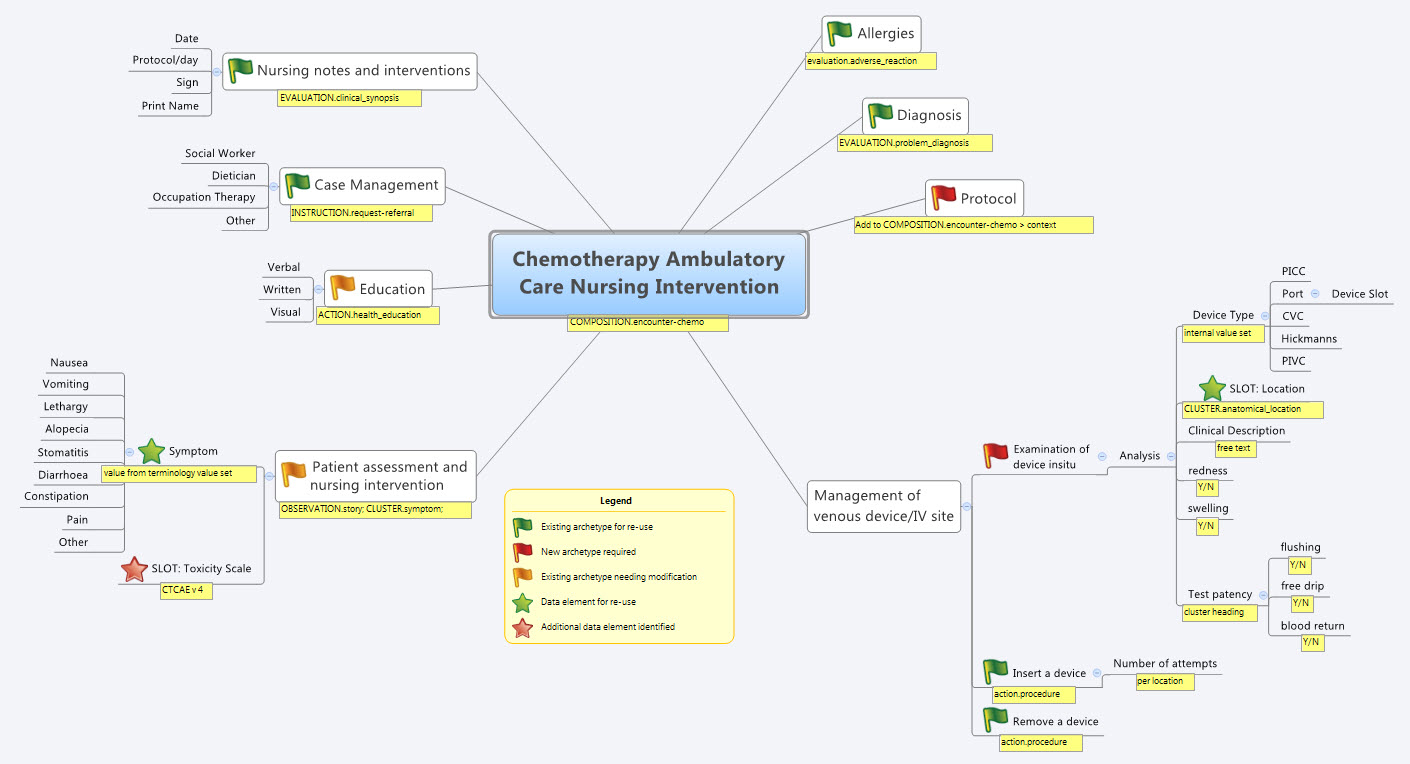
So our practical modelling task was to represent the Chemotherapy Ambulatory Care  Nursing Intervention form. The form had been sourced from another hospital as an example of an existing form and the initial part of the analysis involved working out the intent of the form .
Nursing Intervention form. The form had been sourced from another hospital as an example of an existing form and the initial part of the analysis involved working out the intent of the form .
What I've found over years is that we as human beings are very forgiving when it comes to filling out forms – no matter how bad they are, clinical staff still endeavour to fill them out as best they can, and usually do a pretty amazing job. How successful this is from a data point of view, is a matter for further debate and investigation, I guess. There is no doubt we have to do a better job when we try to represent these forms in electronic format.
We also discussed that this modelling and design process was an opportunity to streamline and refine workflow and records rather than perpetuating outmoded or inappropriate or plain wrong ways of doing things.
So, an outline of the openEHR modelling methodology as we used it:
- Mind map the domain – identify the scope and level of detail required for modelling (in this case, representing just one paper form)
- Identify:
- existing archetypes ready for re-use;
- existing archetypes requiring modification or specialisation; and
- new archetypes needing development
- Specialise existing archetypes – in this case COMPOSITION.encounter to COMPOSITION.encounter-chemo with the addition of the Protocol/Cycle/Day of Cycle to the context
- Modify existing archetypes – in this case we identified a new requirement for a SLOT to contain CTCAE archetypes (identical to the SLOT added to the EVALUATION.problem_diagnosis archetype for the same purpose). Now in a formal operational sense, we should specialise (and thus validly extend) the archetype for our local use, and submit a request to the governing body for our additional requirements to be added to the governed archetype as a backwards compatible revision.
- Build new archetypes – in this case, an OBSERVATION for recording the state of the inserted intravenous access device. Don't take too much notice of the content – we didn't nail this content as correct by any means, but it was enough for use as an exercise to understand how to transfer our collective mind map thoughts directly to the Archetype Editor.
- Build the template.
So by the end of the second day, the trainee modellers had worked through a real-life use-case including extended discussions about how to approach and analyse the data, and with their own hands were using the clinical modelling tooling to modify the existing, and create new, archetypes to suit their specific clinical purpose.
What surprised me, even after all this time and experience, was that even in a relatively 'new' domain, we already had the bulk of the archetypes defined and available in the NEHTA CKM. It just underlines the fact that standardised and clinically verified core clinical content can be re-used effectively time and time again in multiple clinical contexts.The only area in our modelling that was missing, in terms of content, was how to represent the nurses assessment of the IV device before administering chemo and that was not oncology specific but will be a universal nursing activity in any specialty or domain.
So what were we able to re-use from the NEHTA CKM?
- COMPOSITION.encounter
- EVALUATION.adverse_reaction – one instance per adverse reaction included in a adverse reaction list
- EVALUATION.clinical_synopsis
- EVALUATION.problem_diagnosis – one instance per diagnosis included in a problem list
- INSTRUCTION.request-referral – one instance per referral requested
- ACTION.health_education
- ACTION.procedure – with two instances for different purposes
…and now that we have a use-case we could consider requesting adding the following from the openEHR CKM to the NEHTA instance:
- OBSERVATION.story
- CLUSTER.symptom – with multiple instances for each symptom identified
And the major benefit from this methodology is that each archetype is freely available for use and re-use from a tightly governed library of models. This openEHR approach has been designed to specifically counter the traditional EHR development of locked-in, proprietary vendor data. An example of this problem is well explained in a timely and recent blog - The Stockholm Syndrome and EMRs! It is well worth a read. Increasingly, although not so obvious in the US, there is an increasing momentum and shift towards approaches that avoid health data lock-in and instead enable health information to be preserved, exchanged, aggregated, integrated and analysed in an open and non-proprietary format - this is liquid data; data that can flow.
Clinical Knowledge Repository requirements
I've been hearing quite a lot of discussion recently about Clinical Knowledge Repositories and governance. Everyone has different ideas - ranging from sharing models via a simple subversion folder through to a purpose-built application managing governance of combinations of versioned knowledge assets (information models, terminology reference sets, derived artefacts, supporting documentation etc) in various states of publication. It depends what you want to achieve, I guess. In openEHR it became clear very quickly that we need the latter in order to provide a central resource with governance of cohesive release sets of assets and packages suitable for organisations and vendors to implement.
In our experience it is relatively simple to develop a repository with asset provenance and user management. What is somewhat harder is when you add in processes of collaboration and validation for these knowledge assets - this requires development of review and editorial processes and, ideally, display transparency and accountability on behalf of those managing the knowledge artefacts.
The most difficult scenario reflects meeting the requirements for practical implementation, where governance of configurable groups of various assets is required. In openEHR we have identified the need for cohesive release sets of archetypes, templates and terminology reference sets. This can be very complicated when each of the artefacts are in various states of publication and multiple versions are in use in 'on the ground' implementations. Add to this the need for parallel iso-semantic and/or derived models, supporting documents, and derived outputs in various stages of publication and you can see how quickly chaos can take over.
So, what does the Clinical Knowledge Manager do?
- CKM is an online application based on a digital asset management system to ensure that the models are easily accessed and managed within a strong governance framework.
- Focus:
- Accessible resource - creation of a searchable library or repository of clinical knowledge assets - in practice, a ‘one stop shop’ for EHR clinical content
- Collaboration Portal - for community involvement, and to ensure clinical models that are ‘fit for clinical use’
- Maintenance and governance of all clinical knowledge and related resources
- Processes to ensure:
- Asset management
- uploading, display, and distribution/downloading of all assets
- collaborative review of primary* assets to validate appropriateness for clinical use
- content
- translation
- terminology binding
- publication life cycle and versioning of primary assets
- primary asset provenance, differential and change log
- automatic generation of secondary**/derived assets or, alternatively, upload and versioning when auto generation is not possible
- upload of associated***/related assets
- development of versioned release sets of primary assets for distribution
- identify related assets
- quality assessment of primary assets
- primary asset comparison/differentials including compatibility with existing data
- threaded discussion forum
- flexible search functionality
- coordinate Editorial activity
- share notification of assets to others eg via email, twitter etc
- User management
- Technical management
- Reporting
- Assets
- Users
- Editorial activity support
- Asset management
In current openEHR CKM the assets, as classified above, are:
- *Primary assets:
- Archetypes
- Templates
- Terminology Reference Set
- **Secondary assets:
- Mindmaps
- XML transforms
- plus ability to add transforms to many other formalisms, including CDA
- ***Associated assets:
- Design documents
- References
- Implementation guides
- Sample data
- Operational templates
- plus ability to add others as identified
While CKM is currently openEHR-focused - management of the openEHR artefacts was the original reason for it's development - with some work the same repository management, collaboration/validation and governance principles and processes, identified above, could be applied for any knowledge asset, including all flavors of detailed clinical models and other clinical knowledge assets being developed by CIMI, or HL7 etc. Yes, CKM is a currently a proprietary product, but only because it was the only way to progress the work at the time - business models can always potentially be changed :)
It will be interesting to see how thinking progresses in the CIMI group, and others who are going down this path - such as the HL7 templates registry and the OHT proposed Heart project.
We can keep re-inventing the wheel, take the 'not invented here' point of view or we can explore models to collaborate and enhance work already done.
