My presentation delivered remotely from Australia to the Arctic Conference on Dual-Model based Clinical Decision Support & Knowledge Management in Tromsø, Norway today.
The Archetype 'Elevator Pitch'
Know your data?
Recently I was reminded of some work we did a number of years ago. It involved a large research database, painstakingly collected over 20 years. The data was defined across a number of specialisations within a single clinical domain and represented in 83 data dictionaries stored in an Excel spreadsheet.
Data was collected based on a series of questionnaires, and we were told that successive data custodians had, true to human nature, made slight tweaks and updates to the questionnaires on multiple occasions. The data collected was actually evolving!
The only way to view the data was to open each of the 83 spreadsheets, painstakingly, one by one.
We were engaged to create archetypes to represent both the legacy data and the data that the research organisation wished to standardise to take forward.
So the activity of converting these data dictionaries - firstly to archetypes for each clinical concept, and then representing each data dictionary as a template - resulting in considerable insight into the quality and scope of the data that hadn't been available previously.
For example, the mind map below is an aggregation of the various ways that questions were asked about the topic of smoking.
 Interestingly, what it showed was that no one individual in the organisation had full oversight of the detailed data in all of the data dictionaries.The development of the archetypes effectively provided a cross section of the data focusing on commonality at a clinical concept level and revealed insights into the whole data collection that was a major surprise to the research organisation. It triggered an internal review and major revision of their data.
Interestingly, what it showed was that no one individual in the organisation had full oversight of the detailed data in all of the data dictionaries.The development of the archetypes effectively provided a cross section of the data focusing on commonality at a clinical concept level and revealed insights into the whole data collection that was a major surprise to the research organisation. It triggered an internal review and major revision of their data.
Some of the issues apparent in this mind map are:
- A number of questions have been asked in slightly different ways, but with slight semantic variation, thus creating the old 'apples' vs 'pears' problem when all we wanted was a basket of apples;
- Often the data is abstracted and recorded in categories, rather than recording the actual, valuable raw data which could be used for multiple purposes, not just he purpose of the rigid categories;
- Some questions have 'munged' two questions together with a single True/False answer, resulting in somewhat ambiguous data; and
- Some questions are based on fixed intervals of time.
No doubt you will see other issues or have more variations of your own you could share in your systems.
And we have repeatedly seen a number of our clients undergo this same process, where archetypes help to reveal issues with enormously valuable data that had previously been obscured by spreadsheets and the like. The creation of archetypes and re-use of archetypes as consistent patterns for clinical content has an enormous positive impact on the quality of data that is subsequently collected.
And while harmonisation and pattern re-use within one organisation or project can be hard enough, standardisation between organisations or regions or national programs or even internationally has further challenges. It may take a while to achieve broader harmonisation but the benefits of interoperability will be palpable when we get there.
In the meantime the archetypes are a great way to trigger the necessary conversations between the clinicians, domain experts, organisations, vendors and other interested parties - getting a handle on our data is a human issue that needs dialogue and collaboration to solve.
Archetypes are a great way to get a handle on our data.
Archetypes/DCMs MIA in SDOs
Curious to note that there is very little apparent interest in detailed clinical information models (DCMs) of any brand or flavour in the major Standards Development Organisations (SDO's) - they are effectively Missing In Action when compared to the likes of CDA and IHE profiles. The ISO 13972 DCM specification took a long and tortuous time to travel through the ISO TC 215 processes. Engagement with 13972 during its development, and from what I can observe now it is on the verge of publication has been rather sparse. A further piece of work is now starting in ISO regarding quality criteria for DCMs but it also seems to be struggling to find an audience that understands it, or even cares.
I don't quite understand why the concept of DCMs has not been a big ticket item on the radar of the SDOs for a long time as it is a major missing piece of any standards-based framework. Groups like CIMI are raising awareness, alongside the openEHR work, so momentum is gathering, but for some reason it seems to keep a very understated profile compared to new opportunities like FHIR in HL7.
The work of messages, documents, profiles and terminologies are clearly important for interoperability, but standardisation of clinical content models working closely with terminologies can potentially make the work required to develop messages, documents, and profiles orders of magnitude easier.
Let me test a metaphor on you. Think of each message, document or profile as a sentence and each archetype or DCM as a word, a building block that is one component of each sentence. By focussing on the building a specific sentence, we are working backwards by trying to determine the components, and the outcome is still just that single sentence. However if we start by standardising the words/archetypes, then once they are stable it is relatively simple to construct not only one sentence for a specific purpose, but the potential is a much greater output in which many more additional sentences can be created using a variety of words in different combinations. If we manage the words (archetypes) as core building blocks and get them right, then we allow a multitude of possible sentences (messages/documents/profiles) to proliferate.
The ‘brand’ of archetype/DCM solution does not concern me so much as raising awareness that clinician-led, standardised clinical content is a significant missing and overlooked piece of the international eHealth foundations puzzle.
Archetypes: health data bridges
What do we want for our health data - silos of information models for different purposes or ones that bridge multiple use cases? From a series of emails shared on the HL7 Patient Care email list in the past few days...
Grahame Grieve (FHIR, HL7):
"Heather, you need to keep in mind the difference between FHIR and clinical models: it's not our business to say not to exchange data that people do have because some user in an edge case might not understand it. We define an exchange standard, not a clinical UI standard..."
and
"Heather, do not lose sight of the difference between a clinical standard for what care/records should be, and FHIR, which is an IT standard for how care records are."
My response:
"...I am concerned about developing another standard that you state clearly is only designed for exchange and not for what care records should be. If we are not designing to try to harmonise data requirements for health information exchange, how clinical care records are and how clinical care records should be, then we are building siloes of data structures again, that will require mappings and transforms ad infinitum. I’d hate to see us end up with a standard for exchange that can’t be implemented for persistence"...
If we end up with models for exchange, models representing current data in systems (whether or not they represent good clinical practice and models that are regarded as the roadmap for good data, then what have we got? Three sets of data models that perpetuate the nightmare of non-interoperability.
Our openEHR archetypes are attempting to bridge all of these. Use them in whatever context you choose - messages, document exchange, EHR persistence, CDS, secondary use, aggregation and analysis, querying etc. The 'secret sauce' is the use of a second layer of modelling - the template, that allows the correct expression of the archetype appropriate for the context of use.
Mappings and transformations are acceptable where we don't have any choice, especially with legacy data, but they open us up to vulnerabilities from errors, misinterpretation and ambiguity, concerns re data integrity and possible overt data loss. Given the choice, lets work towards creating high quality data that can be re-used in multiple contexts safely.
Oil & water: research & standards
The world of clinical modelling is exciting, relatively new and most definitely evolving. I have been modelling archetypes for over 8 years, yet each archetype presents a new challenge and often the need to apply my previous experience and clinical knowledge in order to tease out the best way to represent the clinical data. I am still learning from each archetype. And we are still definitely in the very early phases of understanding the requirements for appropriate governance and quality assurance. If I had been able to discern and document the 'recipe', then I would be the author of a best-selling 'archetype cookbook' by now. Unfortunately it is just not that easy. This is not a mature area of knowledge. I think clinical knowledge modellers are predominantly still researchers.
In around 2009 a new work item around Detailed Clinical Models was proposed within ISO. I was nominated as an expert. I tried to contribute. Originally it was targeting publication as an International Standard but this was reduced to an International Specification in mid-development, following ballot feedback from national member bodies. This work has had a somewhat tortuous gestation, but only last week the DCM specification has finally been approved for publication - likely to be available in early 2014. Unfortunately I don't think that it represents a common, much less consensus, view that represents the broad clinical modelling environment. I am neither pleased nor proud of the result.
From my point of view, development of an International Specification (much less the original International Standard) has been a very large step too far, way too fast. It will not be reviewed or revised for a number of years and so, on publication next year, the content will be locked down for a relatively long period of time, whilst the knowledge domain continues to grown and evolve.
Don't misunderstand me - I'm not knocking the standards development process. Where there are well established processes and a chance of consensus amongst parties being achieved we have a great starting point for a standard, and the potential for ongoing engagement and refinement into the future. But...
A standards organisation is NOT the place to conduct research. It is like oil and water - they should be clearly separated. A standards development organisation is a place to consolidate and formalise well established knowledge and/or processes.
Personally, I think it would have been much more valuable first step to investigate and publish a simple ISO Technical Report on the current clinical modelling environment. Who is modelling? What is their approach? What can we learn from each approach that can be shared with others?
Way back in 2011 I started to pull together a list of those we knew to be working in this area, then shared it via Google Docs. I see that others have continued to contribute to this public document. I'm not proposing it as a comparable output, but I would love to see this further developed so the clinical modelling community might enhance and facilitate collaboration and discussion, publish research findings, and propose (and test) approaches for best practice.
The time for formal specifications and standards in the clinical knowledge domain will come. But that time will be when the modelling community have established a mature domain, and have enough experience to determine what 'best practice' means in our clinical knowledge environment.
Watch out for the publication of prEN/ISO/DTS 13972-2, Health informatics - Detailed clinical models, characteristics and processes. It will be interesting to observe how it is taken up and used by the modelling community. Perhaps I will be proven wrong.
With thanks to Thomas Beale (@wolands_cat) for the original insight into why I found the 13972 process so frustrating - that we are indeed still conducting research!
Archetype Re-use
Last Thursday & Friday @hughleslie and I presented a two day training course on openEHR clinical modelling. Introductory training typically starts with a day to provide an overview – the "what, why, how" about openEHR, a demo of the clinical modelling methodology and tooling, followed by setting the context about where and how it is being used around the world. Day Two is usually aimed at putting away the theoretical powerpoints and getting everyone involved - hands on modelling. At the end of Day One I asked the trainees to select something they will need to model in coming months and set it as our challenge for the next day. We talked about the possibility health or discharge summaries – that's pretty easy as we largely have the quite mature content for these and other continuity of care documents. What they actually sent through was an Antineoplastic Drug Administration Checklist, a Chemotherapy Ambulatory Care Nursing Intervention and Antineoplastic Drug Patient Assessment Tool! Sounded rather daunting! Although all very relevant to this group and the content they will have to create for the new oncology EHR they are building.
Perusing the Drug Checklist ifrst - it was easily apparent it going to need template comprising mostly ACTION archetypes but it meant starting with some fairly advanced modelling which wasn't the intent as an initial learning exercise.. The Patient Assessment Tool, primarily a checklist, had its own tricky issues about what to persist sensibly in an EHR. So we decided to leave these two for Day Three or Four or..!
So our practical modelling task was to represent the Chemotherapy Ambulatory Care  Nursing Intervention form. The form had been sourced from another hospital as an example of an existing form and the initial part of the analysis involved working out the intent of the form .
Nursing Intervention form. The form had been sourced from another hospital as an example of an existing form and the initial part of the analysis involved working out the intent of the form .
What I've found over years is that we as human beings are very forgiving when it comes to filling out forms – no matter how bad they are, clinical staff still endeavour to fill them out as best they can, and usually do a pretty amazing job. How successful this is from a data point of view, is a matter for further debate and investigation, I guess. There is no doubt we have to do a better job when we try to represent these forms in electronic format.
We also discussed that this modelling and design process was an opportunity to streamline and refine workflow and records rather than perpetuating outmoded or inappropriate or plain wrong ways of doing things.
So, an outline of the openEHR modelling methodology as we used it:
- Mind map the domain – identify the scope and level of detail required for modelling (in this case, representing just one paper form)
- Identify:
- existing archetypes ready for re-use;
- existing archetypes requiring modification or specialisation; and
- new archetypes needing development
- Specialise existing archetypes – in this case COMPOSITION.encounter to COMPOSITION.encounter-chemo with the addition of the Protocol/Cycle/Day of Cycle to the context
- Modify existing archetypes – in this case we identified a new requirement for a SLOT to contain CTCAE archetypes (identical to the SLOT added to the EVALUATION.problem_diagnosis archetype for the same purpose). Now in a formal operational sense, we should specialise (and thus validly extend) the archetype for our local use, and submit a request to the governing body for our additional requirements to be added to the governed archetype as a backwards compatible revision.
- Build new archetypes – in this case, an OBSERVATION for recording the state of the inserted intravenous access device. Don't take too much notice of the content – we didn't nail this content as correct by any means, but it was enough for use as an exercise to understand how to transfer our collective mind map thoughts directly to the Archetype Editor.
- Build the template.
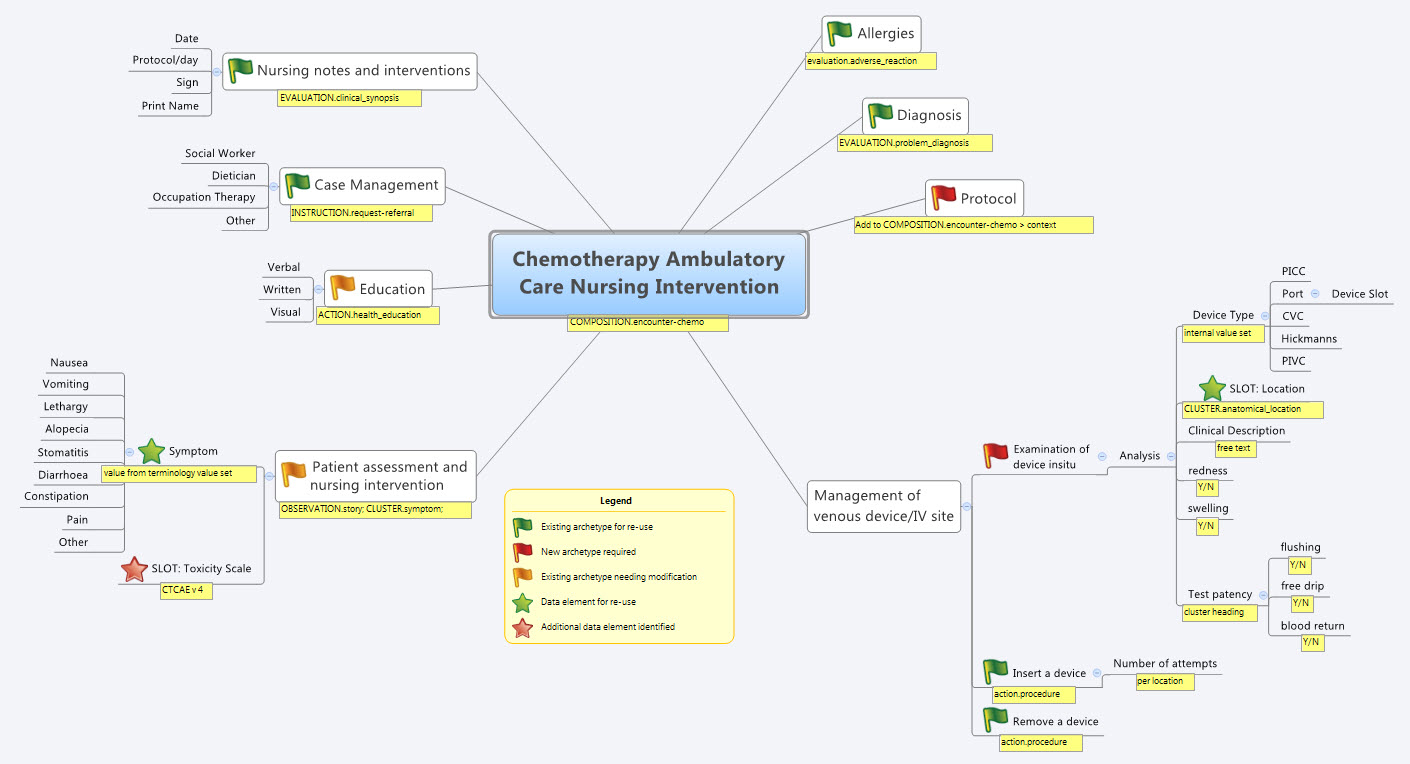
So by the end of the second day, the trainee modellers had worked through a real-life use-case including extended discussions about how to approach and analyse the data, and with their own hands were using the clinical modelling tooling to modify the existing, and create new, archetypes to suit their specific clinical purpose.
What surprised me, even after all this time and experience, was that even in a relatively 'new' domain, we already had the bulk of the archetypes defined and available in the NEHTA CKM. It just underlines the fact that standardised and clinically verified core clinical content can be re-used effectively time and time again in multiple clinical contexts.The only area in our modelling that was missing, in terms of content, was how to represent the nurses assessment of the IV device before administering chemo and that was not oncology specific but will be a universal nursing activity in any specialty or domain.
So what were we able to re-use from the NEHTA CKM?
- COMPOSITION.encounter
- EVALUATION.adverse_reaction – one instance per adverse reaction included in a adverse reaction list
- EVALUATION.clinical_synopsis
- EVALUATION.problem_diagnosis – one instance per diagnosis included in a problem list
- INSTRUCTION.request-referral – one instance per referral requested
- ACTION.health_education
- ACTION.procedure – with two instances for different purposes
…and now that we have a use-case we could consider requesting adding the following from the openEHR CKM to the NEHTA instance:
- OBSERVATION.story
- CLUSTER.symptom – with multiple instances for each symptom identified
And the major benefit from this methodology is that each archetype is freely available for use and re-use from a tightly governed library of models. This openEHR approach has been designed to specifically counter the traditional EHR development of locked-in, proprietary vendor data. An example of this problem is well explained in a timely and recent blog - The Stockholm Syndrome and EMRs! It is well worth a read. Increasingly, although not so obvious in the US, there is an increasing momentum and shift towards approaches that avoid health data lock-in and instead enable health information to be preserved, exchanged, aggregated, integrated and analysed in an open and non-proprietary format - this is liquid data; data that can flow.
Clinical Knowledge Repository requirements
I've been hearing quite a lot of discussion recently about Clinical Knowledge Repositories and governance. Everyone has different ideas - ranging from sharing models via a simple subversion folder through to a purpose-built application managing governance of combinations of versioned knowledge assets (information models, terminology reference sets, derived artefacts, supporting documentation etc) in various states of publication. It depends what you want to achieve, I guess. In openEHR it became clear very quickly that we need the latter in order to provide a central resource with governance of cohesive release sets of assets and packages suitable for organisations and vendors to implement.
In our experience it is relatively simple to develop a repository with asset provenance and user management. What is somewhat harder is when you add in processes of collaboration and validation for these knowledge assets - this requires development of review and editorial processes and, ideally, display transparency and accountability on behalf of those managing the knowledge artefacts.
The most difficult scenario reflects meeting the requirements for practical implementation, where governance of configurable groups of various assets is required. In openEHR we have identified the need for cohesive release sets of archetypes, templates and terminology reference sets. This can be very complicated when each of the artefacts are in various states of publication and multiple versions are in use in 'on the ground' implementations. Add to this the need for parallel iso-semantic and/or derived models, supporting documents, and derived outputs in various stages of publication and you can see how quickly chaos can take over.
So, what does the Clinical Knowledge Manager do?
- CKM is an online application based on a digital asset management system to ensure that the models are easily accessed and managed within a strong governance framework.
- Focus:
- Accessible resource - creation of a searchable library or repository of clinical knowledge assets - in practice, a ‘one stop shop’ for EHR clinical content
- Collaboration Portal - for community involvement, and to ensure clinical models that are ‘fit for clinical use’
- Maintenance and governance of all clinical knowledge and related resources
- Processes to ensure:
- Asset management
- uploading, display, and distribution/downloading of all assets
- collaborative review of primary* assets to validate appropriateness for clinical use
- content
- translation
- terminology binding
- publication life cycle and versioning of primary assets
- primary asset provenance, differential and change log
- automatic generation of secondary**/derived assets or, alternatively, upload and versioning when auto generation is not possible
- upload of associated***/related assets
- development of versioned release sets of primary assets for distribution
- identify related assets
- quality assessment of primary assets
- primary asset comparison/differentials including compatibility with existing data
- threaded discussion forum
- flexible search functionality
- coordinate Editorial activity
- share notification of assets to others eg via email, twitter etc
- User management
- Technical management
- Reporting
- Assets
- Users
- Editorial activity support
- Asset management
In current openEHR CKM the assets, as classified above, are:
- *Primary assets:
- Archetypes
- Templates
- Terminology Reference Set
- **Secondary assets:
- Mindmaps
- XML transforms
- plus ability to add transforms to many other formalisms, including CDA
- ***Associated assets:
- Design documents
- References
- Implementation guides
- Sample data
- Operational templates
- plus ability to add others as identified
While CKM is currently openEHR-focused - management of the openEHR artefacts was the original reason for it's development - with some work the same repository management, collaboration/validation and governance principles and processes, identified above, could be applied for any knowledge asset, including all flavors of detailed clinical models and other clinical knowledge assets being developed by CIMI, or HL7 etc. Yes, CKM is a currently a proprietary product, but only because it was the only way to progress the work at the time - business models can always potentially be changed :)
It will be interesting to see how thinking progresses in the CIMI group, and others who are going down this path - such as the HL7 templates registry and the OHT proposed Heart project.
We can keep re-inventing the wheel, take the 'not invented here' point of view or we can explore models to collaborate and enhance work already done.
Why the buzz about CIMI?
With the recent public statement from the Clinical Information Modelling Initiative (CIMI) my cynical heart feels a little flutter of excitement. Maybe, just maybe, we are on the brink of a significant disruption in eHealth. Personally I have found that the concept of standardising clinical content to be compelling and hence my choice to become involved in development of archetypes. During my openEHR journey over the past 5 or so years it has been very interesting to watch the changing attitudes internationally - from curiosity and 'odd one out' through to "well, maybe there's something in this after all".
And now we have the CIMI announcement...
So what has been achieved? What should we celebrate and why?
At worst, we have had a line drawn in the sand: a prominent group of thought leaders in the international health informatics domain have gathered and, through a somewhat feisty process, recognised that a collaborative approach to the development of a single logical clinical content representation (the CIMI core reference model) is a desirable basis for interoperability across formalisms. Despite most of the participants having significant investment and loyalty to their own current methodology and flavor of clinical models, they have cast aside the usual 'not invented here' shackle and identified a common approach to an initial modelling formalism from which other models will be derived or developed. Whether any common clinical content models are eventually built or not, naming of ADL 1.5 and the openEHR constraint model as the initial formalism is a significant recognition of the longstanding work of the openEHR Foundation team - the early specifications emerged nearly 20 years ago.
At its idealistic best, it potentially opens up a new chapter for health informatics, one that deviates from the relatively safe path of incremental innovation that we have followed for so many years - the reliance on messages/documents/hubs to enable us to exchange health information. There is an opportunity to take a divergent path, a potentially transformational innovation, where the focus is on the data itself, and the message/document/EHR becomes more simply just the receptacle or vehicle for the data. It could give us a very real opportunity to store lifelong health information; simplify data exchange (whether by messages or documents), aggregation, querying and analysis; and support knowledge-based activities such as decision support - all because we will (hopefully) have non-proprietary, common, agreed and fully defined models of clinical content and known transformations between each formalism.
Progress during the next few months will be telling. In January 2012, immediately before the next HL7 meeting in San Antonio, the group will gather again to discuss next steps.
There is a very real risk that despite best intentions all of this will fade away to nothing. The list of participating organisations, including high profile standards organisations and national eHealth programs, is a veritable Who's Who of international health IT royalty, so they will all come with their own (organisational and individual) work experience, existing modelling resources, hope, enthusiasm, cynicism, political agendas, bias and alliances. It could be enough to sink the work of this fledgling group.
But many are battle-weary, having been trudging down this eHealth path for a long time - some now gradually realising that the glacial incremental innovation is not delivering the long-term sustainable answers required for creating 21st Century EHRs as they had once hoped. So maybe this could be the trigger to make CIMI fly!
I think that CIMI is a very bright spark on the health IT horizon. Let's hope that with the right management and governance it can be agilely nurtured into a major positive force for change. And in the future, when its governance is mature and processes robust, we can integrate CIMI into the formal standards processes.
Best of luck, CIMI. We're watching!
CIMI - initial public statement
The following public statement has been released by the Clinical Information Modelling Initiative today:
Public releaseThe Clinical Information Modeling Initiative is an international collaboration that is dedicated to providing a common format for detailed specifications for the representation of health information content so that semantically interoperable information may be created and shared in health records, messages and documents. CIMI has been holding meetings in various locations around the world since July, 2011. All funding and resources for these meetings have been provided by the participants. At its most recent meeting in London, 29 November - 1 December 2011, the group agreed on the following principles and approach.
Principles
- CIMI specifications will be freely available to all. The initial use cases will focus on the requirements of organisations involved in providing, funding, monitoring or governing healthcare and to providers of healthcare IT and healthcare IT standards as well as to national eHealth programs, professional organisations, health providers and clinical system developers.
- CIMI is committed to making these specifications available in a number of formats, beginning with the Archetype Definition Language (ADL) from the openEHR Foundation (ISO 13606.2) and the Unified Modeling Language (UML) from the Object Management Group (OMG) with the intent that the users of these specifications can convert them into their local formats.
- CIMI is committed to transparency in its work product and process.
Approach
- ADL 1.5 will be the initial formalism for representing clinical models in the repository.
- CIMI will use the openEHR constraint model (Archetype Object Model:AOM).
- Modifications will be required and will be delivered by CIMI members on a frequent basis.
- A set of UML stereotypes, XMI specifications and transformations will be concurrently developed using UML 2.0 and OCL as the constraint language.
- A Work Plan for how the AOM and target reference models will be maintained and updated will be developed and approved by the end of January 2012.
- Lessons learned from the development and implementation of the HL7 Clinical Statement Pattern and HL7 RIM as well as from the Entry models of 13606, openEHR and the SMART (Substitutable Medical Apps, Reusable Technologies) initiative will inform baseline inputs into this process.
- A plan for establishing a repository to maintain these models will continue to be developed by the group at its meeting in January.
Representatives from the following organizations participated in the construction of this statement of principles and plan:
- B2i Healthcare www.B2international.com
- Cambio Healthcare Systems www.cambio.se
- Canada Health Infoway/Inforoute Santé Canada www.infoway-inforoute.ca
- CDISC www.cdisc.org
- Electronic Record Services www.e-recordservices.eu
- EN 13606 Association www.en13606.org
- GE Healthcare www.gehealthcare.com
- HL7 www.hl7.org
- IHTSDO www.ihtsdo.org
- Intermountain Healthcare www.ihc.com
- JP Systems www.jpsys.com
- Kaiser Permanente www.kp.org
- Mayo Clinic www.mayoclinic.com
- MOH Holdings Singapore www.mohh.com.sg
- National Institutes of Health (USA) www.nih.gov
- NHS Connecting for Health www.connectingforhealth.nhs.uk
- Ocean Informatics www.oceaninformatics.com
- OpenEHR www.openehr.rog
- Results4Care www.results4care.nl
- SMART www.smartplatforms.org
- South Korea Yonsei University www.yonsei.ac.kr/eng
- Tolven www.tolven.org
- Veterans Health Administration (USA) www.va.gov/health
Further Information:
In the future CIMI will provide information publicly on the Internet. For immediate further information, contact Stan Huff (stan.huff@imail.org)
The DCM environment - a crowdsourcing initiative
There are a number of detailed clinical models available to choose from, from diverse backgrounds, for many differing purposes and with varying degrees of success and penetration. More are arriving on the scene as time goes on – only this week the Clinical Information Models from ONC in the US have been released for their first 2 week period of public comment, and I'd never heard of them before. I haven’t been able to find a single document pointing to an overview on available models – current or past. So, given the current activity development of a quality standard around detailed clinical models in ISO TC 215 and the number of models under development, I thought it timely to try to co-ordinate sourcing a picture of the detailed clinical model landscape.
Well aware that my understanding is limited to the openEHR archetypes I work with, I thought I’d take this opportunity to draw together some of my research and request some assistance from others to crowdsource a picture of the detailed clinical model environment that we operate within.
I have published my initial attempt as a Google doc, see below. The existing information needs to be verified; there are clearly many gaps that need to be filled in; and there are probably other DCMs to be added.
EDIT the DCM environment document...
Anyone can participate, and if you edit, please add your name/org/email to the Contributor list, so that attribution can be made.
Please feel free to edit; to add new DCMs; to correct and verify the facts. Add a column, suggest changes to structure where you see a need... Just make a comment, if you like.
You can clearly see all the gaps in our collective knowledge at present…
Let’s see if we can actually pull together a useful resource. The intent is collaboration to achieve a goal that is possibly greater than most of us can achieve by ourselves.
All contributions are gratefully received and can be freely shared with anyone who is interested.
The latest crowdsourced version of the document appears below. You will need to scroll to seen the entire content.
[googleapps domain="docs" dir="document/pub" query="id=11xGZs4drfwb7TY15Iwpk52UIxZcFhqMcMp1i3yV4To8&embedded=true" /]
________________________________________
Further Comment: 11 August 2011
I've been asked where this work is going and what is planned...
The answer is that I haven’t a specific plan.
To my knowledge there has been no environmental scan to explore what the range of models is, to my knowledge, so thought I’d try myself. I spent a day to develop the initial framework, sparse though it was. Googling was a very frustrating experience and finding out the information that we’re interested in from available websites was not easy.
Hence I thought I’d experiment with the crowd sourcing approach. I know there is a rising tide of interest in the generic concept of detailed clinical models and a lot of confusion about the work in ISO and HL7.
I hoped it might contribute to clarifying the situation and providing a concrete anchor on which to base discussions on detailed clinical models.
Worst case scenario, the final result may be little more than a brainstorming, however it will at least form the basis for anyone interested to launch a more rigorous research program, including potentially feeding into the ISO work. Best case scenario, we may have a significant resource. Final outcome will probably be somewhere in between.
I’m happy to leave it online as a general resource – it seems to have generated a lot of interest. I'm hopeful that it will keep evolving.
Anatomy of a Procedure
ACTION archetypes are one of the harder concepts to grasp in the openEHR clinical modeling world. ACTIONs appear to be promising the world - allowing for recording all activities from planning, scheduling, postponing, cancelling, suspending, through to carrying out and completing an activity. While they may appear cumbersome at first, they are surprisingly elegant and fit for purpose... once you can twist your brain around them.
Consider ACTION archetypes as a walrus - awkward on land, but graceful in the water - well maybe the analogy is being stretched a little, but you get my drift.
Whatever you do, don't underestimate the power of these little clinical models. ACTIONs are designed to enable sophisticated tracking of activities being carried out in a distributed environment - perfect for managing care pathways between multiple providers in disparate locations - not an easy task.
Take a look at this slide show - my attempt to provide a concrete example of how a Procedure ACTION archetype will support documentation about how an INSTRUCTION or order for a Procedure (any procedure, potentially) is carried out in a shared electronic health record environment.
- 'Pathway steps' are identified as the steps (not necessarily sequential) or clinical activities in which it makes sense to record some information in the health record.
- The Data Elements that are identified in the 'Description' include any and all information that we may wish to record for any of the Pathway steps. For example, the data we need to record when we plan to perform a Procedure or the information required to record that the Procedure was performed or abandoned, including the reason for abandonment.
[slideshare id=8563247&doc=201107instructionsactions-110711064154-phpapp02]
Updated: August 17, 2011
Modelling pregnancy
Step 2: Map clinical concepts to archetypes
Map them to existing archetypes or determine which new ones we need. Determine the state of the existing archetypes – do we need to modify or enhance with additional requirements? Consider all possible use cases for each archetype – the end result will be better if we can be as inclusive as possible for all clinical scenarios.
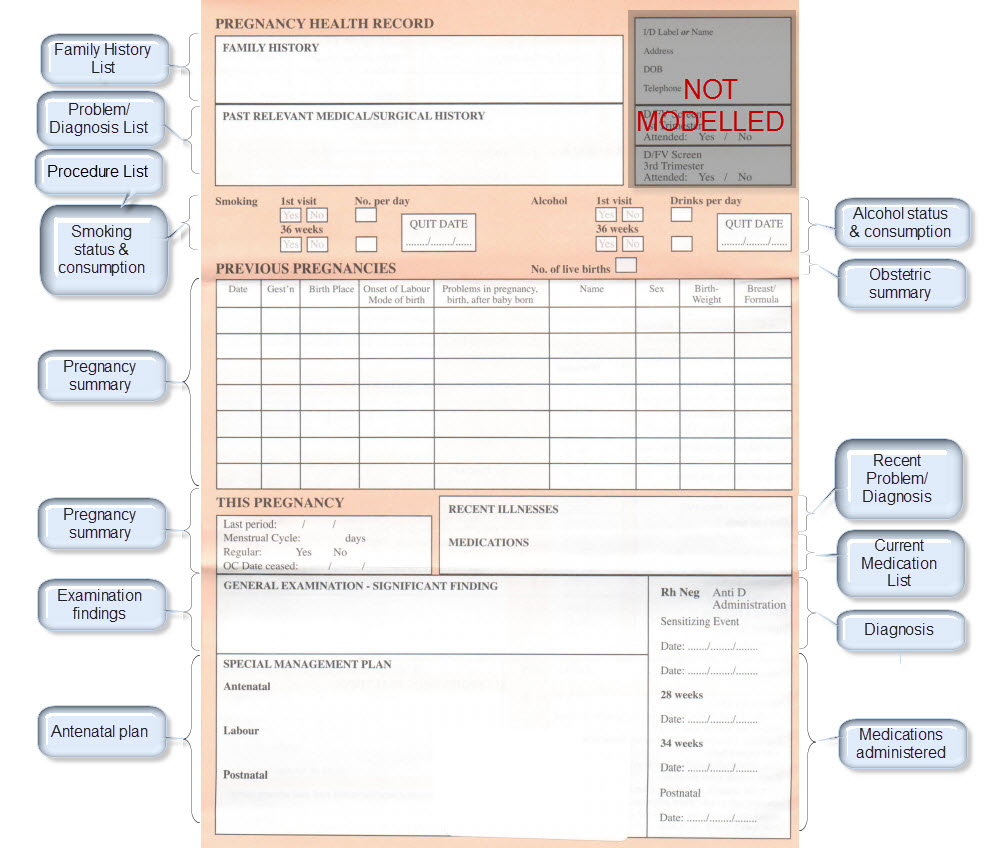
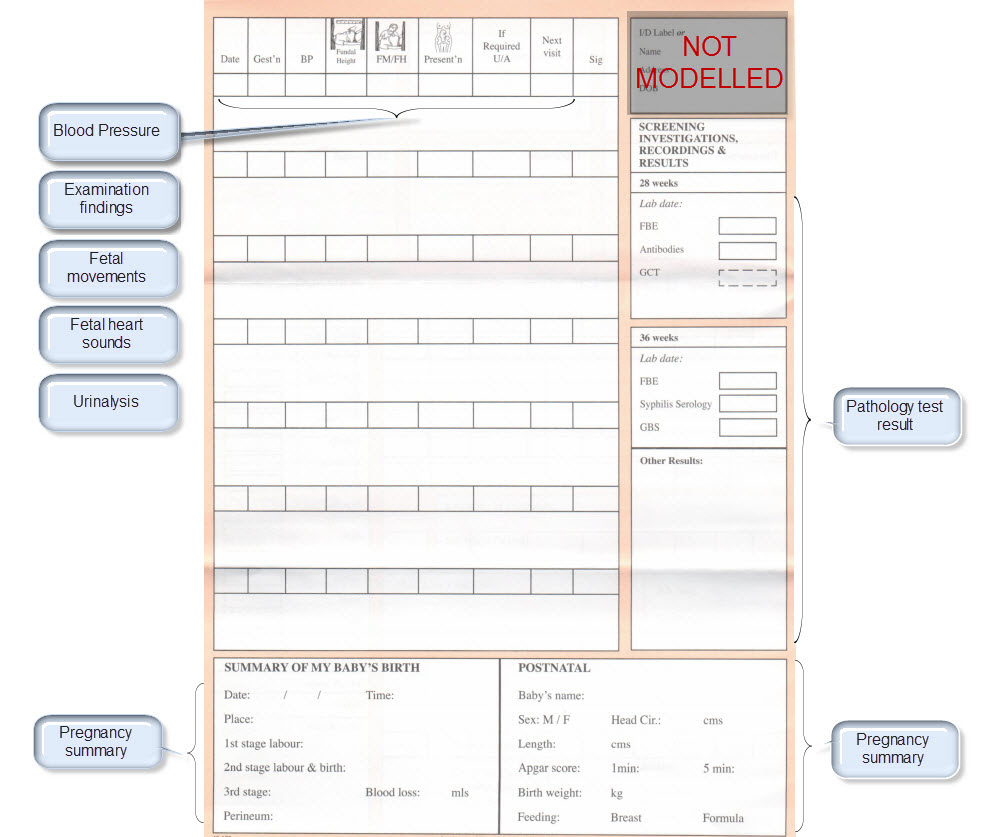
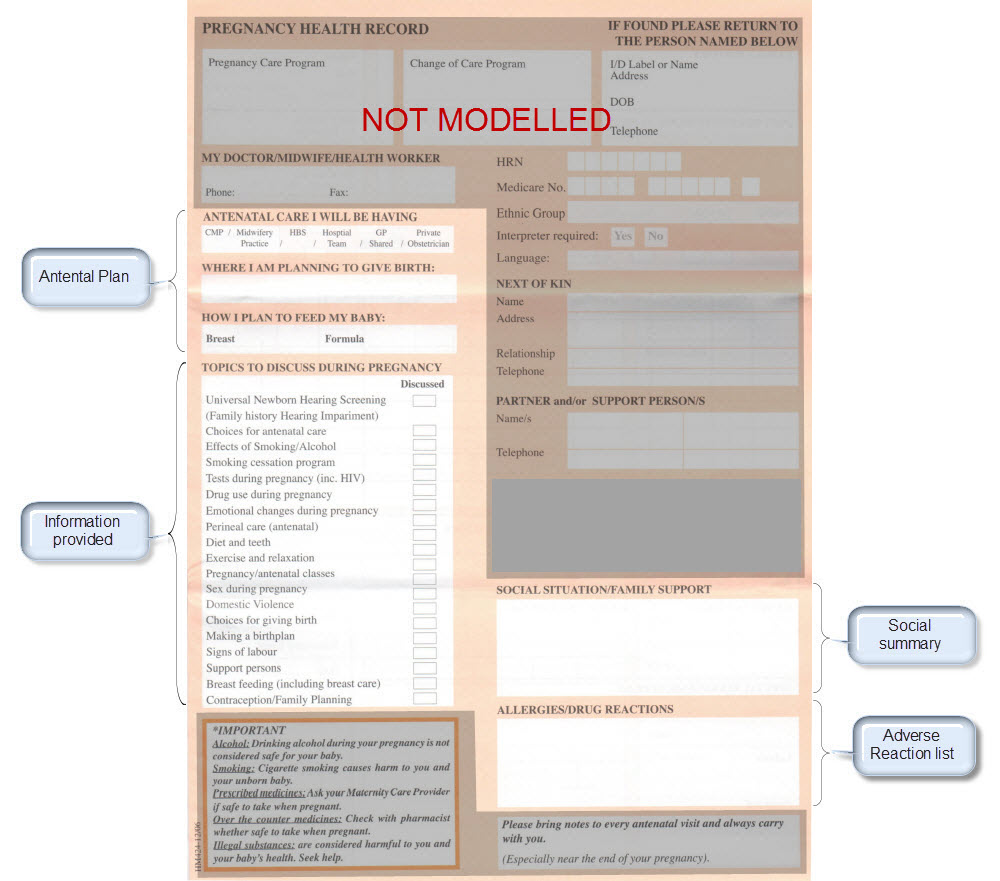
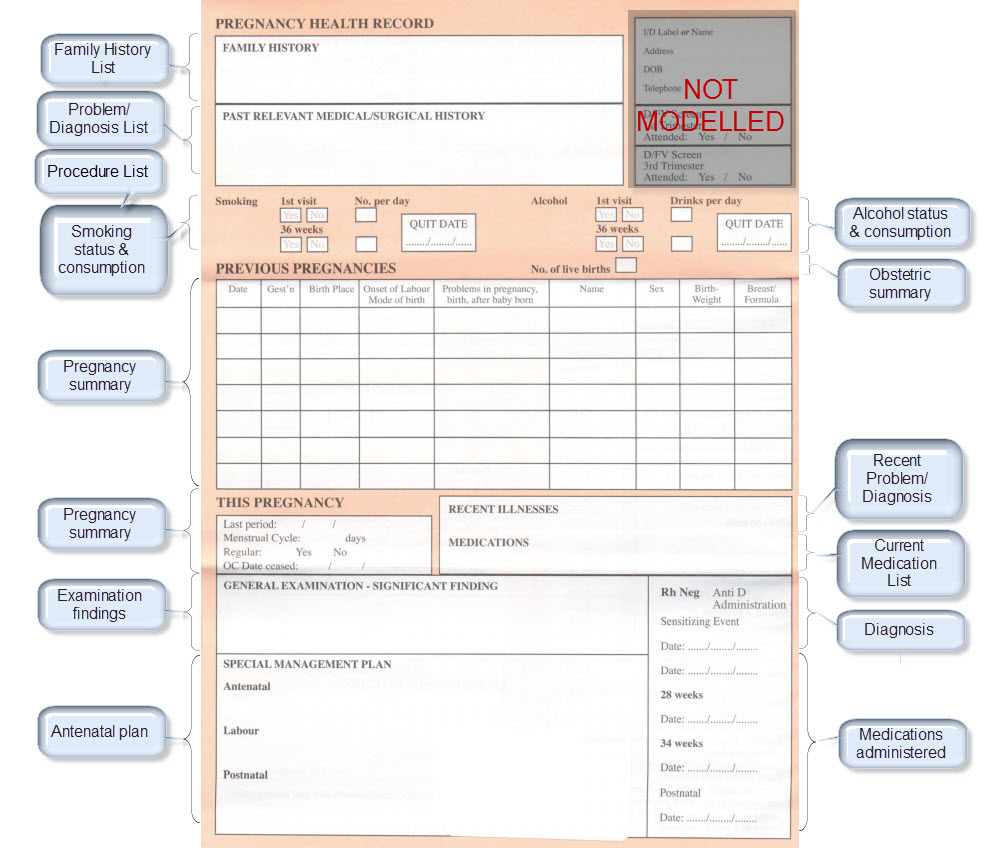
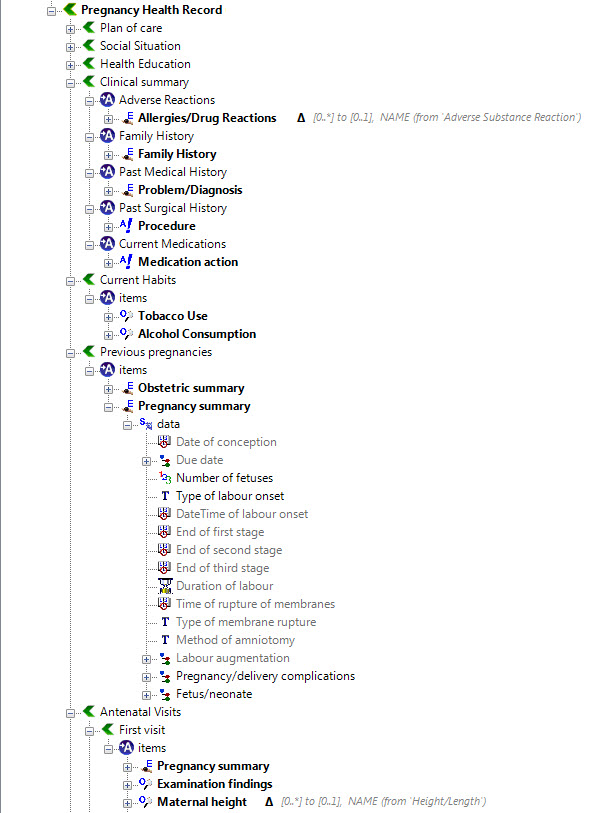
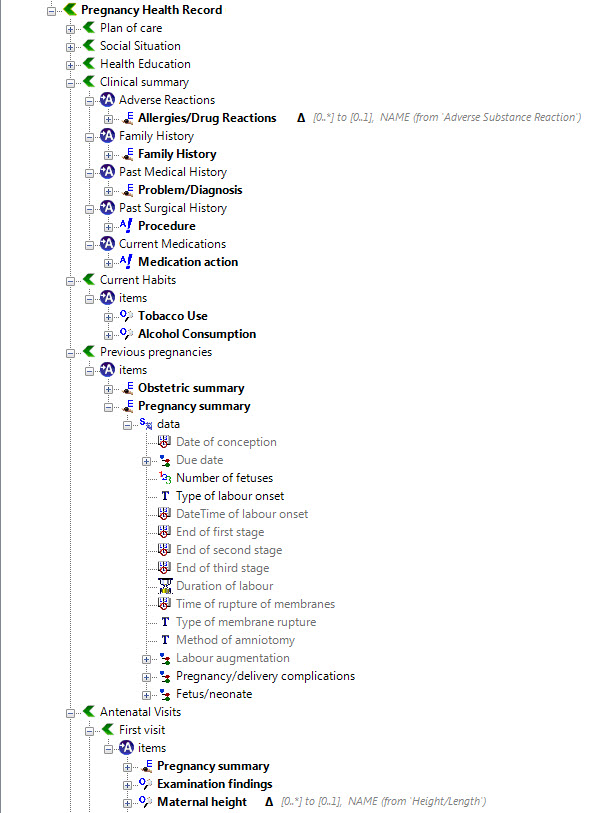
The archetypes identified for this Pregnancy record are:
| Clinical Concept | Comment |
Corresponding archetype concept name NOTE: is linked to the actual archetype in a CKM instance |
| Antenatal Plan | An overarching plan for the patient's antenatal care | Antenatal Plan <INSTRUCTION.antenatal_plan> |
| Information Provided | Details of each piece of health education information discussed or provided to patient | Information Provided <ACTION.health_education> |
| Social Summary | Social Summary <EVALUATION.social_summary> | |
| Adverse Reaction List | Adverse Reaction list is made up of multiple Adverse Reaction entries | Adverse Reaction <EVALUATION.adverse_reaction> |
| Family History List | Family History list is made up of multiple Family History entries | Family History <EVALUATION.family_history> |
| Diagnosis | Problem lists, whether current or past are made up of multiple Problem/Diagnosis entries – the same data pattern will be used in all instances | Problem/Diagnosis <EVALUATION.problem_diagnosis> |
| Problem/Diagnosis List | ||
| Recent Problem/Diagnosis List | ||
| Procedure list | Procedure list is made up of multiple Procedure entries | Procedure <ACTION.procedure> |
| Smoking consumption | Tobacco use <OBSERVATION.substance_use-tobacco> | |
| Alcohol consumption | Alcohol consumption <OBSERVATION.substance_use-alcohol> | |
| Obstetric Summary | Obstetric summary <EVALUATION.obstetric_summary> | |
| Pregnancy Summary | An evolving summary of key data about a single pregnancy – either current or past | Pregnancy summary <EVALUATION.pregnancy_summary> |
| Examination Findings | Examination Findings <OBSERVATION.exam> Examination of the uterus <CLUSTER.exam-uterus> Examination of the fetus <CLUSTER.exam-fetus> | |
| Current Medication List | Medication list is made up of multiple Medication instruction entries | Medication instruction <INSTRUCTION.medication> |
| Medication administered | Medication action <ACTION.medication> | |
| Height | Height/Length <OBSERVATION.height> | |
| Weight | Body weight <OBSERVATION.body_weight> | |
| Body Mass Index | Body Mass Index <OBSERVATION.body_mass_index> | |
| Blood Pressure | Blood Pressure <OBSERVATION.blood_pressure> | |
| Fetal Movements | Fetal Movements <OBSERVATION.fetal_movements> | |
| Fetal heart sounds | Heart rate and rhythm <OBSERVATION.heart_rate> | |
| Urinalysis | The pattern for urinalysis is identical to the pathology test results, and in some cases will be performed in the lab, so the same pattern will be used | Pathology test result <OBSERVATION.pathology_test> |
| Pathology Test results | ||
| Ultrasound results | Imaging examination result <OBSERVATION.imaging_exam> |
KEY: Pink archetype names indicate those that are early drafts. Green archetype names indicate those that are reasonably mature – through previous reviews within CKM Red archetype names indicate those yet to be developed.
Step 3: CKM Review
Within CKM each archetype will undergo review rounds by a range of clinicians, terminologists, software engineers, informaticians and other interested experts. The purpose of each review round will be to fine-tune each archetype so that it meets the requirements for each stakeholder group.
CKM Videos:
Step 4: Create an openEHR template
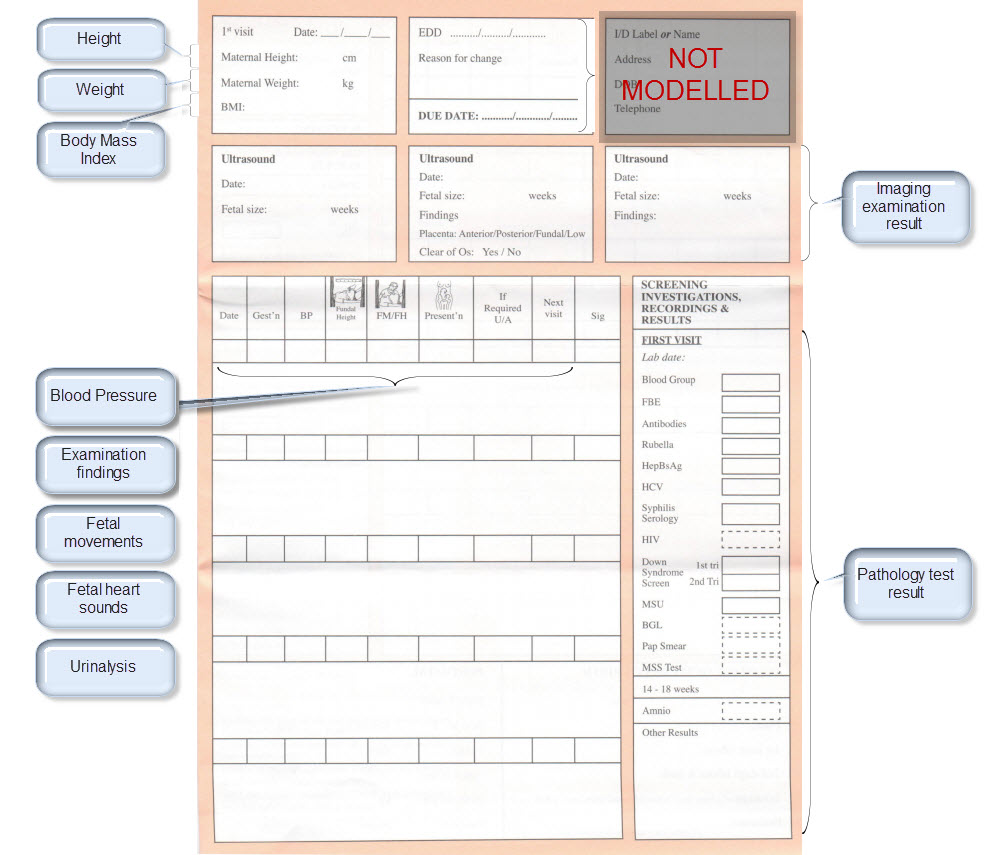
Once community consensus is reached within CKM, the archetypes will be aggregated and constrained in a template to accurately meet the specific use-case requirements for this particular Pregnancy Health Record. Each bold, black heading in the the following screenshot represents an archetype. The Pregnancy Summary archetype has been opened up to see the details. Black text indicates data elements that will be utilised in this use case; grey text indicates inactive data elements.
Other groups may use the same archetypes aggregated in a different order and constrained in a different way to represent their version of the Pregnancy Health Record. As the data in both Pregnancy Records is being recorded according to the same pattern, dictated by the underlying archetype, the information can be exchanged without ambiguity or loss of meaning.
Quality indicators & the wisdom of crowds
Archetype quality I
Up until recently clinical content models, such as archetypes, have been regarded as a novelty; watched from the sidelines with interest from many but not regarded as mainstream. However now that they are increasingly being adopted by jurisdictions and used in real systems, modellers need to change their approach to include processes, methodologies and quality criteria that ensure that the models are robust, credible and fit for purpose. There has been some work done identifying quality criteria for clinical models but there is no doubt that establishing quality criteria for clinical content models is still very much in its infancy:
- Kalra D, Tapuria A, Freriks G, Mennerat F, Devlies J (2008). Management and maintenance policies for EHR interoperability resources [36 pages] (Q-REC Project IST 027370 3.3 ). The European Commission: Brussels. (Last accessed May 28, 2011)
- There has been some slowly progressing work in ISO TC 215 - ISO 13972 Health Informatics: Detailed Clinical Models. Recently it has been split into two separate components, not yet publicly available:
- Part 1. Quality processes regarding detailed clinical model development, governance, publishing and maintenance; and
- Part II - Quality attributes of detailed clinical models
Most of the work on quality of clinical models has been based largely on theory, with few groups having practical experience in developing and managing collections of clinical models, other than in local implementations.
In 2007, Ocean Informatics participated in a significant pilot project. The recommendations were published in the NHS CFH Pilot Study 2007 Summary Report. My own analysis, conducted in December 2007, revealed that there were 691 archetypes within the NHS repository. Of these, 570 were archetypes for unique clinical concepts, with the remainder reflecting multiple versions of the same concept. In fact, for 90 unique concepts there were 207 archetypes that needed rationalisation – most of these had only two versions however one archetype was represented in five versions! We needed better processes!
Towards the end of 2007 a small team within Ocean commenced building an online tool, the Clinical Knowledge Manager to:
- function as a clinical knowledge repository for openEHR archetypes and templates and, later, terminology subsets;
- manage the life-cycle of registered artefacts, especially the archetype content – from draft, through team review to published, deprecated and rejected. Also terminology binding and language translations;
- governance of the artefacts.
In July 2008 we started uploading archetypes to the openEHR CKM, including many of the best from the NHS pilot project. Over the following months we added archetypes and templates; recruited users; and started archetype reviews. All activity was voluntary – both from reviewers and editors. Progress has thus been slower than we would have liked and somewhat episodic but provided early evidence that a transparent, crowd sourced verification of the archetypes was achievable.
In early 2010, Sweden's Clinical Knowledge Manager had its first archetypes uploaded.
In November 2010, a NEHTA instance of the CKM was launched, supporting Australia's development of Detailed Clinical Models for the national eHealth priorities. This is where most collaborative activity is occurring internationally at present.
In this context, I have pondered the issues around clinical knowledge governance now for a number of years, and gradually our team has developed considerable insight into clinical knowledge governance – the requirements, solutions and thorny issues. To be perfectly honest, the more we delve into knowledge governance, the more complicated we realise it to be – the challenge and the journey continues; a lot is yet to be solved :)
It is relatively easy to identify the high level processes in the development of clinical knowledge artifacts, each of which requires identification of quality criteria and measurable indicators to ensure that the final artifacts are fit for purpose and safe to use in our EHR systems. The process is similar for both archetypes and templates; plus the Requirements gathering and Analysis components are applicable to any single overarching project as well.
For archetypes:
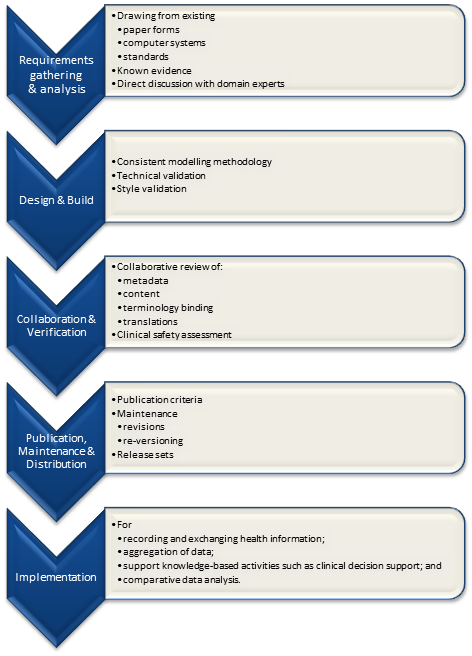
The harder task is that for each of these steps, there are multiple quality criteria that need to be determined, and for each criterion it will be necessary to be able to assess and/or measure them through identifiable quality indicators.
Ideally a quality indicator is a measurement or fact about the clinical model. In some situations it will be necessary to include additional assessments manually performed by qualified experts.
If an indicator can be automatically derived from the Clinical Knowledge Manager (CKM), it ensures that up-to-date assessments of the models are instantly available as the models evolve (such as this Blood Pressure archetype example), and more importantly, without reliance on manual human intervention. However while assessments that do need to be assessed by an expert human – for example, compliance to existing specifications or standards – add valuable depth and richness to the overall quality assessment, they also add a vulnerability due to the need for skilled human resources to not only conduct the assessment, but to apply consistent methodologies during the assessment; these will be much more difficult to sustain.
Assessment of whether the indicators actually satisfy the quality criteria should also ideally be as objective as possible, however our reality is that it will probably more often be subjective and vary depending on the nature of the archetype concept itself. The process cannot be automated, nor can there be a single set of indicators or criteria that will determine the quality of every archetype. We need to ensure appropriate oversight to archetype development, ensuring that a quality process has actually been followed and utilise quality indicators to determine if the quality criteria have been met - on an archetype by archetype basis.
Engaging Clinicians in Clinical Content (in Sarajevo)
Just browsing and found the link to our presentation for a paper the CKM team gave at the Medical Informatics Europe conference in Bosnia, 2009. Thought I'd share it here, in memory of an amazing conference and location:
MIE09: Engaging Clinicians in Clinical Content [PDF]
Our presentation:
[slideshare id=1958920&doc=engagingcliniciansinclinicalcontent-090906091020-phpapp02]
And the reference to Herding Cats is explained (a little) in the embedded video - actually an advertisement for EDS:
[youtube http://www.youtube.com/watch?v=Pk7yqlTMvp8&w=425&h=349]
I arrived after a 37 hour flight - Melbourne - London - Vienna - Sarajevo to join my Ocean CKM team - Ian McNicoll and Sebastian Garde. We presented our paper and also ran an openEHR workshop.
The conference was held at the Holiday Inn in Sarajevo - the hotel in which all the international journalists were holed up during the war, on 'Sniper Alley'.
Despite it being 14 years after the war had ended, raw emotions were palpable many times during the conference. That a pan-European conference was being held in his beloved Sarajevo under his auspice was a very overwhelming event for the Professor of Informatics and he was often seen in tears!
From my hotel room window in the centre of the city I could see 6 cemeteries.
Walking through the inner city we came across many cemeteries, thousands of marble gravestones with the majority representing the young men aged between 18 & 26.
Bullet holes were still very obvious on the walls of buildings.
Yet the city was vibrant, alive and welcoming.
I won't ever forget this visit.
Some photos of the experience:
DCMs – clarifying the confusion
Detailed clinical models are certainly a buzz term in the health IT community in recent years, commonly abbreviated to DCMs. Many people are talking about them but unfortunately, often they are referring to different things. The level of confusion is at least as large as the hype.I sincerely hope that this post helps to lift the veil of confusion just a little...
Anatomy of a Problem... a Diagnosis...
Anatomy of an Adverse Reaction
 Discussion about how to represent some of the commonest clinical concepts in an electronic health record or message has been raging for years. There has been no clear consensus. One of those tricky ones - so ubiquitous that everyone wants to have an opinion - is how to create a computable specification for an adverse reaction.
In the past few years I have been involved in much research and many discussions with colleagues around the world about how to represent an adverse reaction in a detailed clinical model. I'd like to share some of these learnings...
Discussion about how to represent some of the commonest clinical concepts in an electronic health record or message has been raging for years. There has been no clear consensus. One of those tricky ones - so ubiquitous that everyone wants to have an opinion - is how to create a computable specification for an adverse reaction.
In the past few years I have been involved in much research and many discussions with colleagues around the world about how to represent an adverse reaction in a detailed clinical model. I'd like to share some of these learnings...