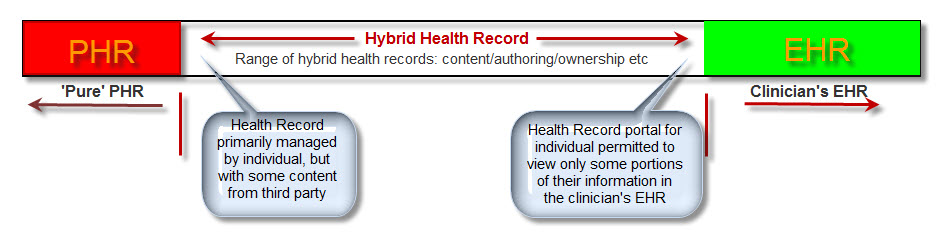
The principles remain unchanged. I've updated some links, and indicated some minor parts that are a little out-of-date. My previous posts, What on earth is openEHR & Connect with openEHR provide up-to-date stats and links.
In a world where connectivity reigns, our health information is largely still caught up in silos and, in the main, is not shareable by clinicians. Shared electronic health records (SEHRs) are increasingly needed to provide timely, comprehensive and coordinated healthcare. Over many years there have been ongoing and thorough attempts to achieve the sharing of health information in order to support the improvement of health outcomes, but this incremental approach, gradually building on previous experience, has not been wholly successful. Progress has been made, but despite enormous investment and resources, the solution has been more difficult than most ever anticipated. Healthcare provision does not seem to fit into the same kind of data sharing model as has been successful in other domains such as banking or financial services.
In order to support interoperability of health information SEHRs need clinical content to be standardised. This assists the move to standardised communication of complex health information between systems and the opening of these vertical domain and organisational silos, allowing accurate and semantically computable health information flow. Inevitably the shared clinical content specifications begin to influence the information models within clinical applications. openEHR is an electronic health record architecture based on many years of research and development, and designed to work in partnership with all vendor systems, organisations and providers to facilitate semantic interoperability of health information. It is a comprehensive and transformational solution which applies to the capturing and sharing of information from the most complex and dynamic knowledge domain – health.
Why are Health Records so difficult?
The interoperability of health information problem is twofold – firstly, the evolving clinical needs and requirements for a SEHR are difficult to pin down, and secondly the technical issues related to a SEHR solution.
Why are clinical requirements so difficult to capture and transform into an electronic health record (EHR)? Certainly no-one will argue that our experience of healthcare delivery is changing – hospital in the home projects; supported self-management; patient requests for access to GP and hospital clinical records; personal health records; service coordination; clinical care plans; preventive health priorities... The list goes on, mirroring the rapidly evolving change in clinical EHR requirements needed to support these new paradigms of healthcare delivery. These new ways of delivering care require a coordinated approach – healthcare providers need to collaborate efficiently in real time, which is not adequately supported by traditional methods such as meetings and phone calls. We really need an interoperable and integrated patient record in which all healthcare providers can participate, within a framework providing governance, authorisation and security measures.
Clinical knowledge domain is very complex and dynamic, so any clinical system created using traditional software development methods (embedding the clinical knowledge directly into proprietary application code and databases) has an uphill battle to deal with the changes. Considering the time it takes for a clinical application to be developed, it may already be out-of-date at its launch! The inherent difficulties in updating knowledge structures that are hard-coded into the application and database inevitably lead to a gradual deterioration of the integrity of the system.
Clinical decision support is another difficult area – there is a lot of discussion about it, but there is very little real computerised decision support happening in practice. Certainly, each clinical system has some that is custom built – usually limited to allergy and drug interaction alerts and a reminder system – but there has been very little general progress in this area. Why is clinical decision support (CDS) still largely found only in the academic domain and not implemented in production clinical systems? We already have clinical guidelines and other resources which are approved and used in paper formats – why are they not integrated in our clinical desktop applications? The answer is multi-factorial. Things that have blocked progress include ownership and sharing of intellectual property, licensing problems etc, but the main issue is to do with practical implementation and cost. An approved guideline can be transformed into some proprietary computable format, but then it has to be able to integrate into every other clinical system – lots of interfaces have to be built, paid for and maintained. This is a nightmare. Another approach is for each system developer to take a guideline and integrate it into their system as best they can. This introduces variability and the potential for significant quality and liability issues to arise.
Silos of proprietary information cannot talk to each other without mappings and necessary assumptions. Such transformations inevitably compromise the quality and integrity of the information being shared, thus creating more threats to patient safety. It is only through use of a common logical clinical content model reflected in our EHRs and CDS systems that the guidelines can be created once according to this agreed model and can be integrated meaningfully and accurately with the clinical information systems to give us quality clinical decision support at the point of care.
From a technical point of view, the current reality around the world is that the majority of vendors are competing to build the greatest and ‘best of breed’ clinical software applications, but all are doing it in their own proprietary way. The software may have a rich functionality and a great user interface. It may do a great job in the clinic, hospital or network on which it is installed, but how does it share clinical data? Some software applications may be able to share with the same system in another geographical location because they share a common data structure, but cannot easily share with the system of another vendor.
Current clinical applications can usually send or receive point-to-point messages using a format which is known and agreed, and based on a standard such as the Health Level 7 (HL7) version 2. The software may also incorporate a terminology to assist in capturing and storing common clinical concepts. However, there is a common misconception that if we simply have a messaging standard paired with a terminology, then we have ‘achieved interoperability’.
A New Paradigm for Electronic Health Records
Most of us mistake the software program for the electronic health record; in fact in the USA the word EHR means a clinical application. The openEHR approach asserts that it is not the application, but rather the data, or health information, which makes up the health record. And further, that it is a common and agreed structure of that data is what makes the electronic health record of any use for computing in health.
In order for two clinical systems to be able to share health data unambiguously, such that clinicians can read it and the computer can compute with it, more is required. This semantic, or knowledge-level, interoperability, based on a common and coherent clinical model, is an absolute requirement for truly shareable EHRs and valuable complementary functionality such as clinical decision support and workflow/care planning. Further, it is only when this clinical content structure is agreed at a local, regional, national or international level, that true semantic interoperability can occur at each of these levels. The broader the level of clinical content model agreement, the broader the potential for semantic health information exchange.
While an increasing number of health informatics experts agree on this view of the problem, incremental work towards EHR standards based on messaging paradigms, such as HL7 v3, have only had limited degrees of success. At an international level, there is increasing concern that this incremental approach to SEHRs may not be able to solve the issues of semantically interoperable EHRs. Remember that Einstein argued: “We can't solve problems by using the same kind of thinking we used when we created them.”
We need semantic interoperability for our SEHRs and in order to achieve this we need a transformational change. We need to use a different paradigm. The openEHR paradigm has been collaboratively developed, reviewed and refined to realise a collective vision of a high quality, internationally interoperable EHR.
What is semantic interoperability?
Let’s be clear about what semantic interoperability means. According to Walker et al[1], Level 4 interoperability, or ‘machine interpretable data’, comprises both structured messages and standardised content/coded data. In practice, it means that data can be transmitted and viewed by clinical systems without need for further interpretation or translation. This is a basic foundation for a truly shareable EHR and for functionality such as clinical decision support.
For clarity, existing HL7 v2 messages probably fit into Level 3 of Walker’s conceptual framework – ‘Machine organisable data’ which comprises structured messages but unstructured content. According to Walker, a level 3 message “requires interfaces that can translate incoming data from the sending organization’s vocabulary to the receiving organization’s vocabulary; usually results in imperfect translations because of vocabularies’ incompatible levels of detail.” We cannot directly compute on a Level 3 message – it requires interpretation or transformation before the computer can use it. HL7 v3 goes a step further with an underlying semantic model. The problem has been that using this in systems has proved very difficult. The HL7 CDA, chosen by Nehta for communication in Australia, bypasses some of the difficulties by at least enabling us to share documents that we can read. More is needed to achieve semantic interoperability.
What is openEHR?
openEHR[2] is a set of open specifications for an Electronic Health Record (EHR) architecture – but it is not a software application. Its design purpose is to enable semantic interoperability of health information between, and within, EHR systems – all in a non-proprietary format, avoiding vendor lock-in of data.
All clinical knowledge concepts are captured in a structured way - known as archetypes – outside the software. The types of archetypes support the recording required for common clinical activities, with some of the key building block archetypes comprising observations, evaluations, instructions and actions. Data built according to these are stored in an EHR in larger ‘composition’ structures, which have their own archetypes. Compositions are comparable to a document that results from a clinical event which is performed e.g. a consultation record or a discharge summary. Archetypes can be simple, such as temperature, blood pressure or diagnosis, or complex, such as capturing the risk to a fetus if the father has a grandmother with Huntingdon’s chorea. The archetypes contain a maximum data set about each clinical concept, including attendant data required such as: protocol, or method of measurement; related events; and context that is required for the clinical data to be interpreted accurately. The creation of archetypes and templates is almost purely a task for clinicians – openEHR archetypes put clinicians in the driver’s seat, enabling them to create the breadth, depth and complexity of the health record to suit their needs for direct healthcare provision.
Aggregations of archetypes are combined in openEHR ‘templates’ in order to capture the data-set corresponding to a particular clinical task, such as an ICU discharge summary or antenatal visit record. When clinicians look at templates, the information contained within them inherently makes sense and doesn’t require significant training for interested clinicians to be able to create templates for their own purposes – be it domain, organisation or purpose specific. Templates can be used to build generic forms to represent the approximate layout of the EHR in a practical sense, and these can be used by vendors to contribute to their user interface development.
Both archetypes and templates can be linked to terminologies or contextually appropriate terminology subsets that will support appropriate term selection by healthcare providers at the point of data entry.
openEHR is rapidly gaining international momentum, supported by evidence from current high profile implementations of the openEHR specifications such as in the UK NHS Connecting for Health program.
Who is openEHR?
The openEHR Foundation owns the intellectual property of the openEHR architecture specifications. The Foundation is a not-for-profit company, with the founding partners being University College London (CHIME department) and Australian company, Ocean Informatics.
The specifications are the result of over 15 years of research and international implementations, including the Good European Health Record (GEHR), and the collaboration of an international community of people who share a common goal – that of the realisation of clinically comprehensive and interoperable electronic health records to support seamless and high quality patient care.
The registered online community has over 1000 members from 75 countries with many actively participating in debate and contributing to ongoing openEHR development from both technical and clinical perspectives.
What is openEHR used for?
openEHR is a specification for secure, shareable health information and provides a foundation on which to build interoperable, modular software applications. These, in turn, can support distributed clinical workflow, such as care plans.
The openEHR specification can be implemented in a number of ways:
- Scalable EHRs – from Personal Health Records to small/medium/large organisations to regional or state clinical record systems and on through to National e-Health programs;
- Message-based, web-service, middleware applications; and
- Integrating existing clinical systems, including virtual federation of data for research or public health purposes.
However, while it has much in the way of additional functionality, it is important not to lose sight of openEHR’s raison d'être – semantic interoperability, or more simply, a truly shareable Electronic Health Record.
How is openEHR different?
There are many aspects of openEHR which clearly differentiate it from other EHR models:
- Open source initiative
The openEHR specifications are freely available under an open licence.
- Separation of the technical and clinical domains
The openEHR design is markedly different to traditional EHR development - it is a 2 level information model. These 2 levels allow a clear separation of the technical reference (i.e. data) model on which software is based from the clinical knowledge itself. The appeal of this new design is that the technical components of the EHR can be kept quite separate from the dynamic clinical knowledge model. In practice, the technicians manage just the technical aspects, while the clinicians are critical in the development of the clinical archetypes and templates that shape the nature of their EHR.
- Purpose-built EHR
The design of the reference and archetype models support many unique openEHR features required for robust clinical record keeping, clinical business process and medico-legal compliance, such as: distributed versioning and merging of EHR records, including audit trails; a strong history and event model for complex observational information; and archetype-driven semantic querying. openEHR designs supports configurable security, anonymous EHRs, fine-grained standards-based privacy, digital signing, and access auditing.
- Knowledge-enabled – capturing the dynamic and complex nature of health information
Clinicians are able to contribute actively and directly to the development of the clinical knowledge models that underpin their EHRs. Archetypes can be revised and versioned to reflect the rapid and varied changes in health domain knowledge.
- Terminology agnostic
openEHR connects flexibly to any or all terminologies through either archetypes or templates. Terminologies such as SNOMED-CT are leveraged when used alongside archetypes, with the archetype clinical model providing context to minimise the need for post-coordination and complexity. Archetypes and terminologies are not competitive but rather complement each other.
- Semantic querying
Archetypes, in combination with terminologies enable powerful possibilities for semantic querying of repository data – whether for individual usage or for population research. Archetype-based querying enables true longitudinal processing of health data, regardless of the originating system.
- Language independent
There is no language primacy in archetypes; they can begin in any language and be translated to multiple other languages. Translation enables the same archetype to be used in different countries, with data created in one language to be interoperable with systems developed and used in other languages. Archetypes are currently available in English, German, Turkish, Dutch, Swedish, Farsi, Spanish and Portuguese.
- Sustainable reference model – life-long EHRs
The openEHR reference model has been rigorously engineered over the past 15+ years as the foundation for a comprehensive health computing platform. It consists only of generic data types, structures and a small number of generic patterns, resulting in a small, stable and sustainable information model for IT people to maintain. This approach allows a clinical data repository to act as a future-proof data store, totally independent of software applications and technology change. In practice this means that no software application changes, or redeployment, are required when new or revised archetypes are published to reflect changing clinical knowledge. As a result, life-long, application-independent health records are possible for the first time.
- Ease of implementation
openEHR is comparatively easy to implement:
- there is little infrastructure required;
- the software required is small, due to being based on a compact and stable, object-oriented reference model; and
- the clinical models (Archetypes) can be developed separately from the software application.
- Ongoing development and enhancement
Based on feedback from its international collaborators, openEHR is undergoing continuing development, with ongoing maintenance and releases of specifications and software.
- Governance of shared content
Archetypes are created once, and if broadly agreed upon, they can become the basis of consistent sharing of data content between systems, providers and even other countries. While archetypes can be designed and used by a given solo practitioner/researcher or locally within an organisation, the power of archetypes becomes more evident the more broadly they are shared. All systems that use the same archetype – even across country borders and terminologies – will be able to interoperate. The openEHR Foundation is in the process of developing an ontologically based international, open source archetype and template repository[3]. This is being supported by a governance framework which will facilitate archetypes to be submitted for international clinical review, and publication. It comprises a complete archetype and template lifecycle and version management repository and supporting processes to allow countries, regions, organisations or vendors to freely download and utilise these commonly agreed archetypes in their clinical systems. Archetype libraries can also be set up at other levels, as needed – domain, organisational, regional, or national.
- A collaborative model of development rather than a standards-based path
The openEHR development to date has been the result of an interested and motivated volunteers from a broad international community of clinicians and software engineers. Formal and rigorous processes have underpinned its progress and both Architectural and Clinical Review Boards, comprising world experts in their fields, have had oversight of the overarching strategy, process and governance. Standards are not rejected in the openEHR approach; they are just not a part of the development process. In comparison to a traditional standards-based approach, openEHR development has been relatively rapid and pragmatic, rather than being held back by the slower process of ‘design by committee’. In fact, the recent European CEN standard for EHR extracts (EN13606) has been based on an earlier version of, and a subset of, openEHR. In addition, openEHR’s Archetype Definition Language (ADL), has recently been accepted as an International Standards Organisation (ISO) committee draft, for consideration as a standard.
Where is openEHR being used? [as at October, 2007]
openEHR is being used in both active research and commercial activities. [4] Research on openEHR is being conducted in Sweden, Australia, United Kingdom, USA, Sri Lanka and Spain. Commercial development is occurring in Australia, United Kingdom’s NHS Connecting for Health, Netherlands, Belgium, Sweden, Turkey and the USA.
The United Kingdom’s National Health Service (NHS) Connecting for Health program has just commenced a formal clinical modelling program using openEHR archetypes and templates to provide a common and agreed clinical content on which to base its clinical applications. In a pilot early in 2007, content developed for NHS Maternity and Emergency domains were provided to vendors for implementation in new clinical application development.[5] These archetypes are available in the public domain, and have undergone broad internal review by expert clinicians prior to being approved for NHS usage. The Emergency templates developed reflect the top 10 presentations to an Emergency Department – including chest pain, shortness of breath and collapse. The Maternity templates followed the clinical journey of a pregnant woman – from a pre-pregnancy consultation and antenatal visits, through to capturing the labour and delivery record, including Partogram data. Each template is made up of a variable number of archetypes – ranging from a few simple templates containing only 2 or 3 archetypes through to complex templates containing up to 80 discrete clinical concepts.
Current openEHR status and supporting tools [as at October, 2007]
- openEHR Specification
The latest openEHR Specification (Release 1.0.1) was published on 15 April 2007.
- Clinical Content Development of archetypes and templates has gained significant momentum in the past 12 months:
- Archetypes
As a result of initial Australian modelling work and, more recently, within the UK NHS, there are now approximately 250 archetypes.
- Templates
Over forty distinct templates representing clinical needs in Emergency and Maternity were developed over a short time period in March/April 2007.
- Commercial Tools
Currently there are a number of commercial tools supporting the open source openEHR implementation. The major tools to date are developed in .Net and Java. Australia’s Ocean Informatics[6] has developed a suite of products supporting openEHR implementation in .Net, while Sweden’s Linkoping University have developed an Archetype Editor in Java and Cambio Healthcare Systems[7] have an early Java kernel.
The Ocean .Net products include:
- Clinical Knowledge Suite:
-
 Archetype Editor (open source) and Template Designer, including semi-automated screen form construction based on templates;
Archetype Editor (open source) and Template Designer, including semi-automated screen form construction based on templates;
- Terminology Toolset – including a caching terminology server and subsetting tool, incorporating SNOMED-CT in the first instance; and
- Archetype-powered Query Designer
- OceanEHR platform – including a universal data repository, demographic service binding, generic EHR Viewer; and data integration & transformation services.

- Archetype/Template Library Service as an online template and archetype repository
Get involved with openEHR
Membership of the openEHR community is free and open to everyone via the openEHR website[8]. It assumes a commitment to a common vision for high quality, interoperable EHRs, and a willingness to share ideas and experience. All levels of interest and contribution are welcome – from novice to expert. The lists range from discussion on technical, clinical and implementer mailing lists through active development of archetypes, templates and tools, to review and critique of clinical content models and technical specifications. Whether you are a clinician, patient advocate, vendor or provider of health care you have a role in the development of what is becoming known as 'the world's record'.
[1] Walker J, Pan E, Johnston D, Adler-Milstein J, Bates DW, Middleton B. The Value of Health Care Information Exchange and Interoperability. Health Affairs 2005 Jan 19 - http://content.healthaffairs.org/cgi/content/abstract/hlthaff.w5.10v1
[2] For more details, see www.openEHR.org
[3] For more details, see www.archetypes.com.au [UPDATE 2010: Clinical Knowledge Manager - www.openEHR.org/knowledge]
[4] For more details, see www.openehr.org/projects/t_projects.htm [UPDATE 2010: http://www.openehr.org/projects/eiffel.html]
[5] For more details of the NHS clinical modeling work, see www.ehr.chime.ucl.ac.uk/display/nhsmodels/Home
[6] For more details, see www.oceaninformatics.com
[7] For more details, see www.cambio.se/zino.aspx?lan=en-us
[8] www.openehr.org
 Sharing a beer, or three, with Keith Boone. I was chuffed when he blogged that he was coming to meet me! And I’m pleased to report that I seemed to have some impact on him after showing some of our collaborative work happening on the openEHR CKM.
Sharing a beer, or three, with Keith Boone. I was chuffed when he blogged that he was coming to meet me! And I’m pleased to report that I seemed to have some impact on him after showing some of our collaborative work happening on the openEHR CKM.